A FRESH LOOK AT THE MICROBES IN HEALTHY REEF SYSTEMS
by Timothy A. Hovanec, Ph.D.

An excerpt from the September/October 2021 issue of CORAL Magazine
“For every complex problem, there is an answer that is clear, simple… and wrong.” —H.L. Mencken
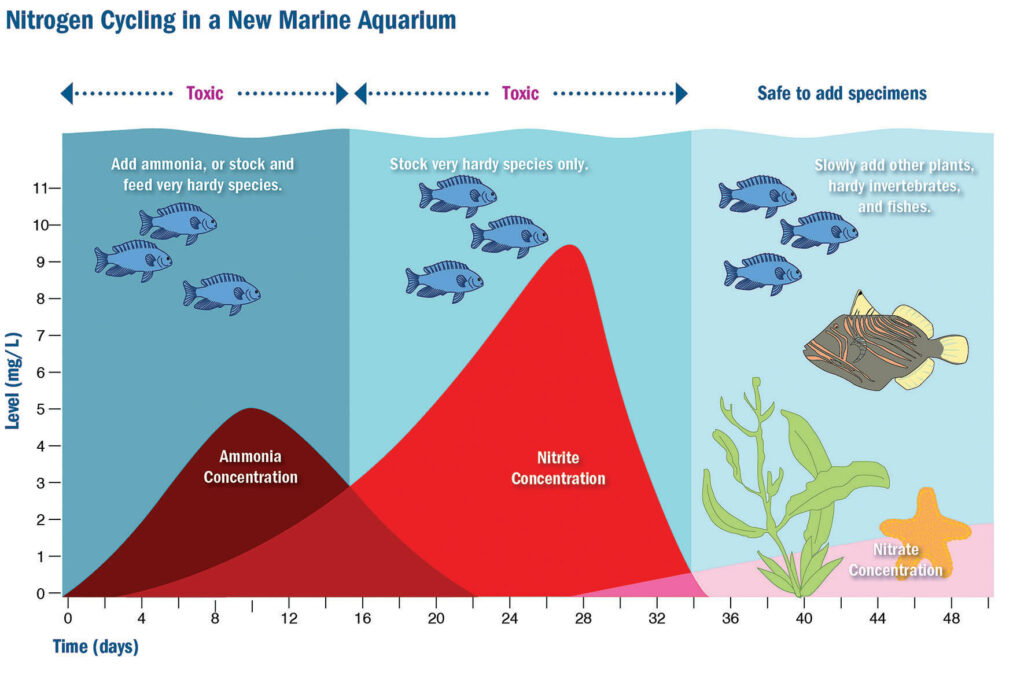
A long, long time ago, when all things seemed simpler, nitrification in the marine aquarium was known as vital for success— but an easy-enough process to understand:
- Ammonia and nitrite in water are toxins originating as animal wastes and uneaten food, able to kill life in an aquatic system. (At high enough levels, a poisoned fish can suffer hyperexcitability, loss of equilibrium, increased respiratory rate, and convulsions, and it may lapse into a coma and die.) In the early days of marine aquarium keeping, it was called “new tank syndrome.”
- Through a process known as the Nitrogen Cycle, ammonia (potentially deadly) is converted to nitrite (less toxic, but still a threat), nitrite is then converted to nitrate (harmless).
Done. Straightforward. Simple. It was known that this nitrification was done by bacteria: Nitrosomonas europaea was the ammonia-oxidizing bacteria (AOB) and the nitrite it produced was converted to nitrate by the nitrite-oxidizing bacteria Nitrobacter winogradskyi (NOB). It didn’t matter whether the aquarium was freshwater, brackish or marine—this was the process and that’s who did it. End of story.
The problem is that everything written above is wrong when it comes to nitrification in aquatic environments—and aquaria in particular. Even though much of what I am about to describe has been known for 15 to 20 years or more, most authors writing in the aquarium literature, even today, gloss over nitrification using a version or variation of the classic, obsolete Nitrogen Cycle scenario. A rewriting of the textbooks and countless digital reference works is long overdue.
Before delving into what we know today, it’s important to get something clear at the start: new research is published seemingly every month in the scientific literature about nitrifying bacteria and nitrification, with results from samples obtained from aquaria and aquaculture systems, and the story of nitrification and nitrifiers is not settled. Here and in the next issue we will present a good general overview of the current knowledge, but some details are bound to change in the near and distant future; that’s how science works.
Consider some of the old mythology and stock answers to basic questions regarding the nitrification process in aquaria and the organisms responsible that still abound today:
- Q: Is nitrification in aquaria done by bacteria?
A: Yes… but…. - Q: Is nitrification a two-step process done by two different organisms?
A: Yes… but…. - Q: Do Nitrosomonas europaea oxidize ammonia and Nitrobacterwinogradskyi oxidize nitrite?
A: Yes… but….
The “yes… but….” answers add complications to the story. Humans like simple organization and linear trends, which is why we classify things and try to rearrange complex patterns, but this is biology, which means it can be messy. The aim of this article is to bring us all up to date with the process of nitrification, name the major players, dispel some myths, and show how understanding the process and the players better can make cycling more efficient, saving you time and money. Let’s start first with: Why? It is important to understand nitrification because:
A. Controlling ammonia, the principal waste product of fish and the major source of nitrogen input to aquaria, is key to balancing your tank’s water quality over the long-term.
B. Microorganisms are key to having a successful aquarium, and nitrifiers can make up a high percent of the microorganisms in your tank.
C. Every tank must go through nitrification (i.e., cycle). Understanding the organisms responsible can save you time, money, and headaches during this period.
D. Fishless cycling is a very popular process to start your tank, and the difference between success and frustration is understanding the process and the bugs.
E. If you decide to use a bacterial starter or booster product, you’ll be better informed on how to proceed.
THE BASICS
The vast majority of marine fish we keep in aquaria are classified as teleosts or bony fish—animals that excrete ammonia as their primary nitrogen waste product. This contrasts with sharks, skates, and rays (elasmobranchs), which produce urea much like humans. Ammonia (as the gaseous form [NH3]) passes through their gills into the water via a process called passive diffusion. This is an energy-saving mechanism—the fish do not have to spend energy converting ammonia to urea or getting rid of a majority of their nitrogenous waste.
Once the ammonia (NH3) enters the aquarium water, it forms its conjugate pair ammonium (NH4 +). Ammonia is considered a base because it can accept hydrogen ions from the water, while ammonium is the conjugate acid to ammonia, as ammonium can release one of its hydrogen ions reforming ammonia. The accepting and releasing of hydrogen goes back and forth constantly.
The pH of the aquarium water is the main factor that determines the percentages of ammonia-ammonium present, with water temperature playing a minor secondary role. At low pH values (6.5 and lower), 99.7 percent or more of the total ammonia is in the ammonium form, while at high pH values (11.5 and higher), 98 percent or more of the total ammonia is in the form of ammonia. For convenience, many authors use “ammonia” to mean all the ammonia present, not just the NH3 faction. Here I will use “total ammonia” when discussing the sum of the ammonia (NH3) and ammonium (NH4 +) factions, to be clearer. Thus:
Total Ammonia = Ammonia (NH3) + Ammonium (NH4 +)
Ammonia (NH3) is a gas, which is why you can smell ammonia in cleaning solutions made with ammonium hydroxide (the pH is >12), but there is no scent of ammonia from ammonium chloride solutions (pH 4.6– 6.0). Further, ammonia is un-ionized, meaning it has no charge, so it is neither a cation (positively [+] charged ion) nor an anion (negatively [-] charged ion). Ammonium (NH4 +) is a cation due to its positive charge.
This distinction is important because ammonia, having no charge, can easily pass through cell membranes, which is what makes ammonia the toxic form of total ammonia. As previously mentioned, ammonia is the form passively excreted by fish, but the direction of flow can be reversed. If the ammonia (note the form) concentration in the aquarium water is higher than the ammonia concentration in the fish, ammonia will flow from the water into the fish and lead to ammonia poisoning.

This presents us with a short side road into practical advice: One of many controversial topics in the marine fishkeeping hobby is acclimating new fish arrivals. Think about what is happening in the bag. The fish are respiring, producing carbon dioxide that lowers the pH of the water, while at the same time excreting ammonia. So, the bag water has high total ammonia but a low pH, which means most of the total ammonia is in the non-toxic ammonium form, so the fish are relatively safe from ammonia poisoning. Now we open the bag and start dripping clean seawater into it. This causes the pH to rise, which shifts more of the total ammonia into the toxic ammonia form and starts to stress the fish. Depending on the length of acclimation time in the bag, you could be doing more harm than good to the fish. While there are always exceptions, in most cases getting the fish out of the bag water quickly and into fresh seawater (without thermal shock, of course) is better for the fish than a slow drip acclimation, at least in this author’s opinion and experience.
As previously stated, ammonia from fish is the main source of nitrogen going into the aquarium. Ammonia (not ammonium) is also the form the nitrifying bacteria utilized, contrary to what most authors write. Again, this is because ammonia (being un-ionized) can flow across the cellular membrane, while ammonium can’t. The oxidation of ammonia to nitrate is a two-step process:
- NH3 + 1.5 O2 -> NO2 + H2O + H+
- NO2- + H2O -> NO3- + 2H+
As the equations show, at each step of the nitrification process hydrogen ions (H+) are produced. The hydrogen ions are initially neutralized by the buffer capacity of your water (the common carbonate buffering system in seawater accepts the H+), but inevitably the pH of the water will drop, due to the constant input of hydrogen ions produced by nitrification. pH measures the concentration of hydrogen ions in the water, and it is a negative number, so adding more hydrogen ions means a less negative number. That is why a pH of 7.8 has more hydrogen ions than a pH of 8.2.
The average pH of the ocean is about 8.1, with minor differences due to depth or location. Marine fish, along with most organisms in the ocean, are not well adapted to changes in pH, because there was no evolutionary reason. This contrasts with freshwater fish, which can experience large pH changes over the course of the year. (This is also why acidification threatens the majority of life in the Earth’s oceans, as organisms from seaweeds to invertebrates to fish are not well adapted to the lower pH values that will result from the seas absorbing the excess carbon dioxide produced by human activities.)
Now that we have a basic understanding of the process, let’s mess it up a little. It has been considered a basic “rule” that nitrification was done by two different organisms: one to convert the ammonia, another to convert the nitrite. In 2015, this was shown not to be the case; a single organism was isolated that could convert ammonia to nitrate, sometimes forming the nitrite intermediate.
Before that, it was demonstrated that ammonia oxidation was not done solely by bacteria but that microorganisms from the Domain Archaea were important ammonia-oxidizing organisms. These newly discovered ammonia-oxidizing archaea (AOA) were, in fact, found and isolated from marine aquaria!
So, let’s rewind a little and go from the process to the players responsible.

THE PLAYERS
Nitrifying bacteria have been known since the 1890s from the efforts of the legendary Russian soil microbiologist Sergio Winogradsky to develop both an understanding of the process and isolate the organisms involved. As was the norm in microbiology until the late 1980s, most of the original work on nitrifiers involved the arduous task of trying to develop pure cultures, which is time-consuming and, in many cases, resulted in failures. However, Winogradsky, as far back as the 1920s, preached working with “environmental” samples rather than pure cultures of single species. But this process still involved manipulating the culture environment to try to simulate the natural environment.
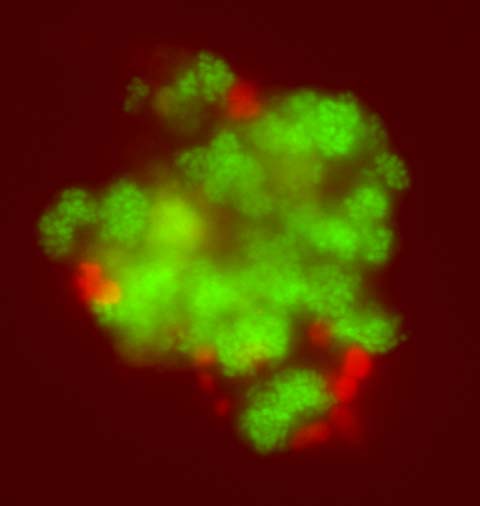
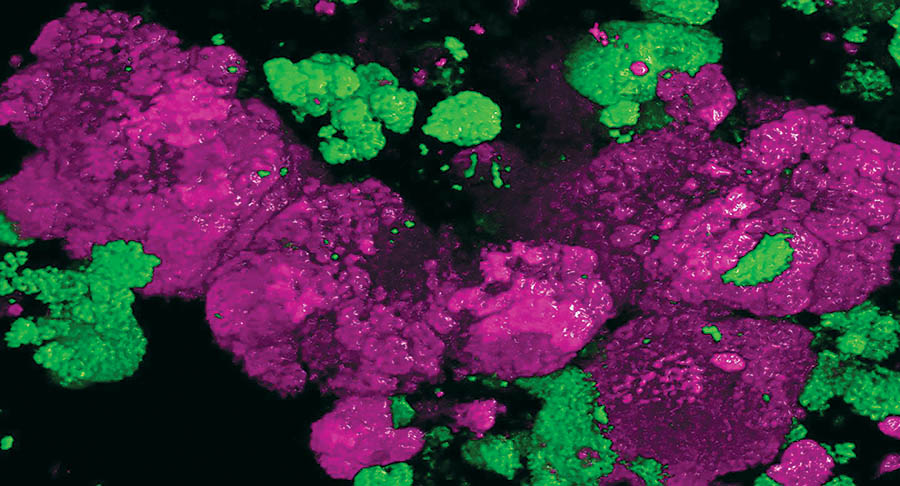
Microbiologists knew that these culture-dependent methods resulted in some level of bias, as the methods would select for organisms that could grow best with the medium or environment provided. But there weren’t yet alternatives, and the results, while not perfect, were assumed to be a good representation of what organisms were most numerous or important in the environment sampled. Nitrifying bacteria are a classic example of this. Thus, the accepted scientific explanation became that ammonia was oxidized (technically losing an electron) to nitrite by a bacterium called Nitrosomonas europaea (Winogradsky 1891 and 1892), and nitrite was further oxidized to nitrate by another bacterium, Nitrobacter winogradskyi, which was officially described in 1917 but known earlier.
This essentially became the story of nitrification. A few other nitrifying organisms were found, but they were generally ruled unimportant because they were from extreme or unusual environments. The reason for this is that in nearly every case the same general culture method was utilized to isolate or at least increase the number of nitrifiers in the target sample. The problem was that the lab culture method favored the growth of Nitrosomonas europaea and Nitrobacter winogradskyi, so that was the answer one got. Whether looking at sewage treatment, soil, or fish ponds, these two bacteria kept showing up as the answer.
There were clues that these culture-dependent methods were inadequate. Johnson and Sieburth in 1976 pointed to this conclusion when they were unable to find species of Nitrobacter in the nitrifying environment of a fish culture system’s biofilter that they examined with transmission electron microscopy. Then the story took another hit when research showed that bottled, commercially available mixtures of nitrifying bacteria used to accelerate the start-up of nitrification either did not work or were of limited use.
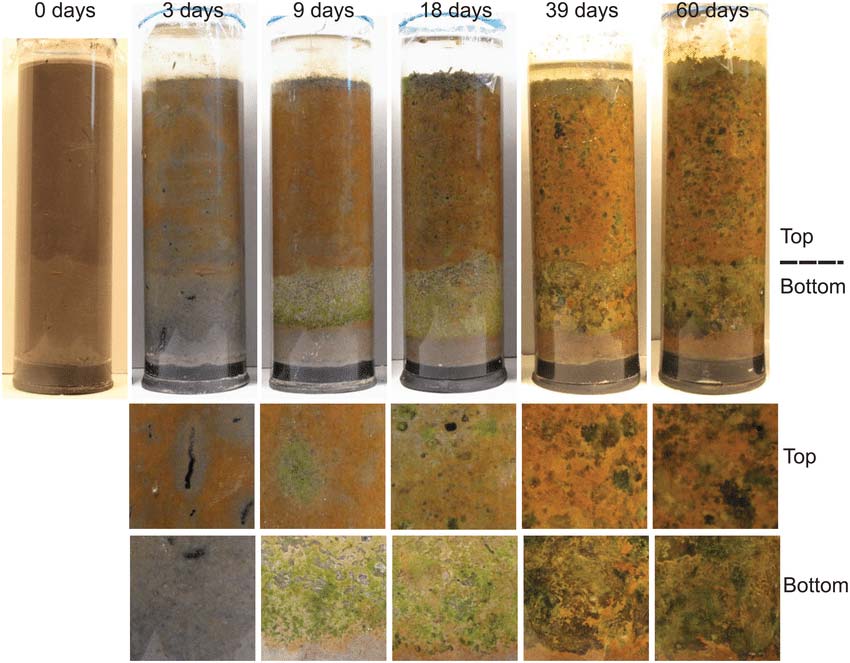
As is common in any good tale, there were a few side plots that one needs to know about. Starting in the late 1970s and exploding in the 1990s, new analytical methods for microbiology and its sister science of microbial ecology (studying microorganisms in their environment) opened up a whole new micro-world and level of understanding.
THE THREE DOMAINS OF LIFE: ENTER ARCHAEA
At the cellular level, what makes a species a species, different from another species, is its genes. More specifically, it’s the DNA and RNA of the genes, or, diving deeper into molecular biology, the sequence of the nucleotides that make up the DNA/RNA (which is as deep as we go in this dive). One ribosomal RNA gene, the 16S, shortened to 16S rRNA, occurs in all self-replicating organisms and holds the key to our story.
Most of us learned in an early biology class there are [were] five Kingdoms of Life: Animals, Plants, Fungi, Protists and Monera, the latter two being simple, single-celled organisms, such as algae, amoebas, and bacteria. However, this classification was not really based on any underlying scientific criteria but more on a thinking that it is easy to tell a plant from an animal—and, for those that can’t easily be thrown into one or the other, we’ll create a few catch-all groups. At a slightly more scientific level, all organisms can be classified as eukaryote (containing a membrane-bound nucleus inside a cell) or prokaryote (lacking a cell nucleus or any membrane-bound organelles in a cell). But, again, this classification is lacking in some scientific respects.
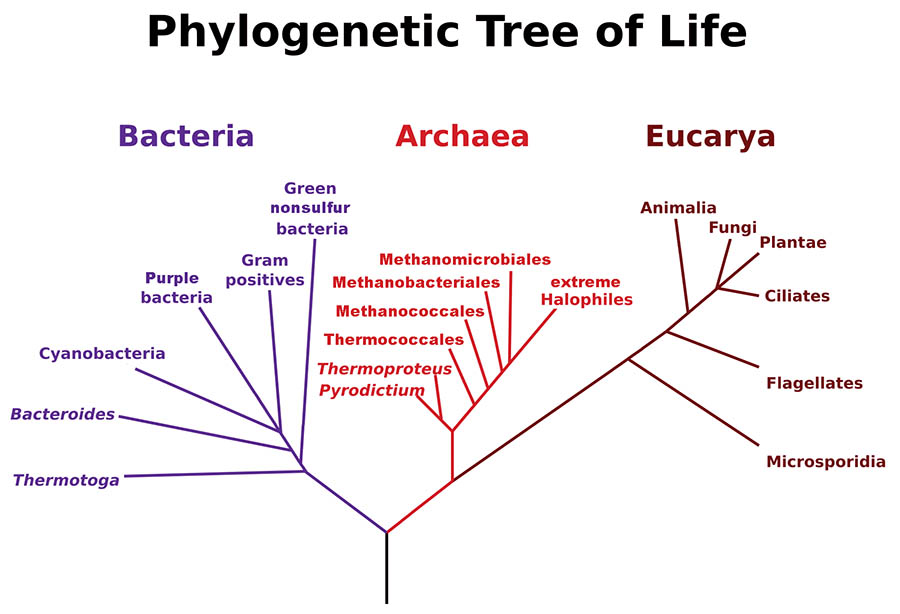
In 1977, a new Three Domain classification system was proposed by American microbiologist Carl Woese. It was based on the nucleic acid sequences of the 16S rRNA gene. This system was based upon the science that every extant organism has this gene, which is not too long and which can be sequenced (yielding phylogenetically-based data). Without getting too technical, there are regions on the gene where the nucleic acid sequence is exactly the same for all organisms, and other regions that are the same for all bacteria, or all eukaryotes and the same sequence for a third group—the Archaea. Thus, Woese proposed a new classification and called the Domains Eubacteria (“true bacteria”), Archaea (single-celled organisms without cell nuclei), and Eukarya (all the higher taxonomic groups, all with membrane-bound organelles and cell nuclei).
Woese’s concept was not immediately accepted, especially the part about Archaea being a separate domain. However, this is not the place to go further into phylogenetics (for brevity and clarity of concept, I am simplifying), but I encourage the curious reader to Google or do a library search for Carl Woese and learn more about the discovery of the Third Domain of Life, which now dominates how we think and classify organisms. Much remains to be learned about Archaea, but it is increasingly clear that they play important roles in the microbiota of different organisms and habitats, including coral reefs.
What is important to understand is that:
- Now the classification of organisms could be done based on genetic information rather than what they look like (which is quite hard for bacteria/archaea, which do not look that much different from each other)
- There is no need to culture the organisms—instead, in situ samples can be taken and processed without any further manipulation (called culture-independent methodology)
- Tools could be developed that target specific regions of the 16S rRNA gene at the genus and species level. One set of these tools is molecular probes.
These three traits combined to break open not only the study of nitrifying bacteria but the entire field of microbial ecology—along with our revised understanding of the role of bacteria and other microbes in the successful startup and operation of marine aquariums.
Continue to Part 2…coming soon!
Editor’s note: In Part II, Dr. Tim Hovanec will describe his own work in isolating the nitrifying microbes active in marine aquarium systems and will discuss how the aquarist can better manage aquarium cycling and maintain healthy populations of beneficial nitrifiers.
Timothy A. Hovanec, Ph.D., is a microbiologist and founder of Dr. Tims Aquatics in Moorpark, California. For 17 years, he was the Chief Science Officer of Aquaria Inc., the parent company of Marineland Aquarium Products, Aquarium Systems (Instant Ocean), and Perfecto Manufacturing. Dr. Hovanec has authored numerous scientific papers in aquatic microbial ecology and in public aquaria and aquaculture fields, and he is a former President of the Pet Industry Joint Advisory Council.
REFERENCES
References
Bagchi S., S.E. Vlaeminck, L.A. Sauder, M. Mosquera, J.D. Neufeld, and N. Boon. 2014. Temporal and Spatial Stability of Ammonia-Oxidizing Archaea and Bacteria in Aquarium Biofilters. PLoS ONE 9(12): e113515. https://doi.org/10.1371/journal.pone.0113515
Bartelme, R.P., S.L. McLellan, and R.J. Newton, 2017. Freshwater recirculating aquaculture system operations drive biofilter bacterial community shifts around a stable nitrifying consortium of ammonia-oxidizing archaea and comammox Nitrospira. Front. Microbiol. 8, https://doi.org/10.3389/fmicb.2017.00101.
Bik HM, Alexiev A, Aulakh SK, et al. 2019. Microbial community succession and nutrient cycling responses following perturbations of experimental saltwater aquaria. mSphere 4: e00043-19. https://doi.org/10.1128/mSphere.00043-19.
Bower, C.E. and D.T. Turner. 1981. Accelerated nitrification in new seawater culture systems: effectiveness of commercial additives and seed media from established systems. Aquaculture 24:1-9.
Bower, C.E. and D.T. Turner. 1984. Evaluation of two commercial nitrification accelerators in closed seater culture systems. Aquaculture 41:155-159.
Brown, M.N. A. Briones, J, Diana and L. Raskin. Ammonia-oxidizing archaea and nitrite-oxidizing nitrospiras in the biofilter of a shrimp recirculating aquaculture system. FEMS Micro. Ecol. doi:10.1111/j.1574-6941.2012.01448.x
Burrell. P.C., J. Keller and L.L. Blackall. 1998. Microbiology of a Nitrite-Oxidizing Bioreactor. Appl. Environ. Microbiol. 64(5): 1878-1883.
Burrell, P.C., C. M. Phalen and T. A. Hovanec. 2001. Identification of Bacteria Responsible for Ammonia Oxidation in Freshwater Aquaria. Appl. Environ. Microbiol. 67(12): 5791-5800.
Daims, H., Lebedeva, E., Pjevac, P. et al. 2015. Complete nitrification by Nitrospira bacteria. Nature 528, 504–509 doi.org/10.1038/nature16461
DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl Acad. Sci. USA 89:5685-5689.
Dworkin, M. 2012. Sergie Winogradsky: a founder of modern microbiology and the first microbial ecologist. FEMS Microbiol. Rev. 36: 364-379. doi:10.1111/j.1574-6976.2011.00299.x
Gieseke, A., U. Purkhold, M. Wagner, R. Amann, and A. Schramm. 2001.Community structure and activity dynamics of nitrifying bacteria in a phosphate-removing biofilm. Appl. Environ. Microbiol. 67:1351–1362.
Hovanec, T.A. 1986. Acute Ammonia Toxicity and Ammonia Excretion of Striped Bass (Morone saxatilis). Master’s Thesis San Diego State Univ., 140p.
Hovanec, T.A. 1998. Characterization of the Nitrifying Bacteria in Aquaria and Mono Lake, California Using Molecular Methods. Ph.D. Dissertation Univ Calif Santa Barbara, 203 p.
Hovanec, T.A. 2001. U.S. Patent No. 6,207,440. Issued March 21, 2001. Bacterial Nitrite Oxidizer.
Hovanec, T.A. 2005. European Patent No. EP 1 282 688. Issued March 16, 2005. Ammonia oxidizing bacteria.
Hovanec, T.A. 2007. U.S. Patent No. 7,267,816. Issued Sept. 11, 2007. Ammonia Oxidizing Bacteria.
Hovanec, T. A. and E. F. DeLong. 1996. Comparative Analysis of Nitrifying Bacteria Associated with Freshwater and Marine Aquaria. Appl. Environ. Microbiol. 62(8): 2888-2896.
Hovanec, T. A., L. T. Taylor, A. Blakis and E. F. DeLong. 1998. Nitrospira-like bacteria associated with nitrite oxidation in freshwater aquaria. Appl. Environ. Microbiol. 64(1): 258-264.
Huang, Z., Y. Jiang, X. Song, E, Hallerman, L. Peng, D. Dong, T. Ma, J. Zhai and W. Li. 2018, Ammonia‑oxidizing bacteria and archaea within biofilters of a commercial recirculating marine aquaculture system. AMB Express 8:17. https://doi.org/10.1186/s13568-018-0551-1
Hüpeden, J., B. Wemheuer, D. Indenbirken, C. Schulz and E. Spieck. 2020. Taxonomic and functional profiling of nitrifying biofilms in freshwater, brackish and marine RAS biofilters. Aqua. Eng. 90:102094
Johnson, P.W. and J.M. Sieburth. 1976. In situ morphology of nitrifying-like bacteria in aquaculture operations. Appl. Environ. Microbiol. 31: 423-432.
Juretschko, S., G. Timmermann, M. Schmid, K.-H. Scheifer, A. Pommerening-Röser, H.-P. Koops and M. Wagner. 1998. Combined Molecular and Conventional Analyses of Nitrifying Bacterium Diversity in Activated Sludge: Nitrosococcus mobilis and Nitrospira-Like Bacteria as Dominant Populations. Appl. Environ. Microbiol. 64:3042-3051
Koch, H., M. A. H. J. van Kessel and S. Lücker. 2019. Complete nitrification: insights into the ecophysiology of comammox Nitrospira. Appl. Micro. Biot. 103:177-189.
Koops, H.-P., U. C. Böttcher, A. Möller, A. Pommerening-Röser, and G. Stehr. 1991. Classification of eight new species of ammonia-oxidizing bacteria: Nitrosomonas communis sp. nov., Nitrosomonas ureae sp. nov., Nitrosomonas aestuarii sp. nov., Nitrosomonas marina sp. nov., Nitrosomonas nitrosa sp. nov., Nitrosomonas eutropha sp. nov., Nitrosomonas oligotropha sp. nov., Nitrosomonas halophila sp. nov. J. Gen. Microbiol. 137:1689–1699.
Könneke, M., A.E. Bernhard, J.R. de la Torre, C.B. Walker, J.B. Waterbury and D.A. Stahl. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543-546.
Liu, S., Wang, H., Chen, L. et al. 2020. Comammox Nitrospira within the Yangtze River continuum: community, biogeography, and ecological drivers. ISME J 14, 2488–2504. https://doi.org/10.1038/s41396-020-0701-8
Noller, H. 2013. Carl Woese: Discoverer of life’s third domain, the Archaea. Nature 493:610.
Oren, A. 1999. Bioenergetic aspects of halophilism. Micro. Mole. Biol. Rev. 63:334-348.
Randall, D.J. and T.K.N. Tsui. 2002. Ammonia toxicity in fish. Mar.Poll.Bull. 45:17-23.
Princic, A., I. Mahne, F. Megusar, E. A. Paul, and J. M. Tiedje. 1998. Effects of pH and oxygen and ammonium concentrations on the community structure of nitrifying bacteria from wastewater. Appl. Environ. Microbiol. 64:3584–3590.
Purkhold, U., A. Pommerening-Roser, S. Juretschko, M. C. Schmid, H.-P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis:implications for molecular diversity surveys. Appl. Environ. Microbiol. 66: 5368–5382.
Schramm, A., D. de Beer, M. Wagner and R. Amann. 1998. Identification and Activities In Situ of Nitrosospira and Nitrospira spp. as Dominant Populations in a Nitrifying Fluidized Bed Reactor. Appl. Environ. Microbiol. 64: 3480–3485.
Strous, M. et al. . 1999. “Missing lithotroph identified as new planctomycete”. Nature 400(6743): 446–449.
Tal, Y., J.E.M. Watts, S.B. Schreier, K.R. Sowers and H.J. Schrier. 2003. Characterization of the microbial community and nitrogen transformation processes associated with moving bed bioreactors in a closed recirculated mariculture system. Aquaculture 215:187-202.
Urakawa, H., Y. Tajima, Y. Numata and Satoshi Tsuneda. 2008. Low Temperature Decreases the Phylogenetic Diversity of Ammonia-Oxidizing Archaea and Bacteria in Aquarium Biofiltration Systems. Appl. Environ. Microbiol. 74(3):894-900.
Urakawa, H. and A.J. Sipos. 2020. Application of the consortia of nitrifying archaea and bacteria for fish transportation may be beneficial for fish trading and aquaculture. Aqua. Res. 51:3429-3442.
Van de Graaf A.A., A. Mulder, P. De Bruijn, M.S.M. Jetten, L.A. Robertson and J.G. Kuenen. 1995. Anaerobic oxidation of ammonium is a biologically mediated process. Appl. Environ. Microbiol. 61(4): 1246–1251. doi:10.1128/AEM.61.4.1246-1251.
Van Kessel, M., Speth, D., Albertsen, M. et al. 2015. Complete nitrification by a single microorganism. Nature 528, 555–559. 2015. https://doi.org/10.1038/nature16459
Winogradsky, S. 1891. Recherches sur les organisms de la nitrification. Ann Inst Pasteur 5: 577–616.
Winogradsky, S. 1892. Contribution á la Morphologie des Organismes de la Nitrification. Arch. Sci. biol. (St Petersburg),1. Abstract in Ann. Inst. Pasteur, 6 , 459.
Woebken, D., P. Lam, M.M.M. Kuypers, S.W.A. Naqvi, B. Kartal, M. Strous, M.S.M. Jetten, B.M. Fuchs, and R. Amann. 2008. A microdiversity study of anammox bacteria reveals novel Candidatus Scalindua phylotype in marine oxygen minimum zones. Environ. Micro. 10 (11):3106-3119. doi:10.1111/j.1462-2920.2008.01640.x.
Woese, C.R. 1994. There Must be a Prokaryote Somewhere: Microbiology’s Search for Itself. Microbiol. And Mol. Biol. Rev. 58(1):1-9.
Woese, C.R. and G.E. Fox. 1977. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc. Natl. Acad. Sci. USA 74(11):5088-5090.







Okay. Can you please take a look at this equation.
NH3 + 1.5 O2 -> NO2 + H2O + H+
NO2- + H2O -> NO3- + 2H+
How did the no2 in the first equation suddenly become an no2- in the second? It seems quite odd (forgive me, I am a high schooler learning chemistry)
And if it is Mr. Matt Pedersen answering me, are any of your Moorish Idols or obligate corallivore butterlfyfsih still alive, and if so. How long have you had them
Brandon, I’ll look into your first question before I address it.
To your OTHER question, the Moorish Idols, along with every last fish in that system, were lost when an unidentified pathogen came into the fishroom. I had two aquarium fish vets examine things, and the suspected culprit was a virus. The result being I had to bleach the entire system, and it sat empty for a couple years. It is currently running with freshwater fish in it. MAYBE someday.
The Redfin X Melapterus Butterflyfish pair is still alive and well. I’d have to go back and look to see how long it’s been, but at least 5-6 years now.
Brandon,
Not sure if you are still looking for an answer to your chemistry question but if you are perhaps this will help.
Count the number of valence electrons for each compound:
NH3 + 1.5 O2 –> NO2 + H2O + H+
8 + 18 –> 17 + 8 + 0
When viewed this way we can see that we are missing exactly one electron on the product side. That idicates that the correct equation would most likely have NO2- (Nitrite) rather than NO2 (Nitrogen Dioxide). My guess is this is simply at typo and it should have been NO2-.
This is what I am thinking as well, but am not going to make the edit until I have it confirmed.
PS personally how do you prefer to maintain a healthy population of bacteria, and what uniquenesses have you observed about the nitrogen cycle in your tank? Have you noticed any way the nitrogen cycle in the tank deviated from the simplified Nitrosomonas-niteobacter format? If the unique ways your nitrogen cycle works caused any problems, how did you fix them
Please, Where can i read part two?
File this in the “We Forgot To Do That” dept. Looks like we never actually posted Part II online! You can read Part II in the November/December 2021 issue of CORAL, but I’ve added publishing Part II online, here, to my to-do list. Can’t promise if or when, but it’s on the list!